Equilibrium¶
examples/equilibrium.py demonstrates how scaling can be used together with tolerances to achieve desired accuracy from the numerical integration.
We will consider the transient towards an equilibrium for a dimerization:
\[\begin{split}A + &B &\overset{k_f}{\underset{k_b}{\rightleftharpoons}} C\end{split}\]
The analytic solution is (its derivation is left as an exercise):
\[\begin{split}A(t) &= A_0 - x(t) \\
B(t) &= B_0 - x(t) \\
C(t) &= C_0 + x(t) \\
x(t) &= \frac{(U-b)(U+b)(e^{Ut}-1)}{2k_f(Ue^{Ut} + U - qe^{Ut} + q)} \\\end{split}\]
where
\[\begin{split}U &= \sqrt{A^2k_f^2 + 2ABk_f^2 - 2Ak_bk_f + B^2k_f^2 -
2Bk_bk_f + 4Ck_bk_f + k_b^2} \\
q &= Ak_f + Bk_f - k_b\end{split}\]
$ python equilibrium.py --help
usage: equilibrium.py [-h] [--tend TEND] [-A A0] [-B B0] [-C C0] [--nt NT]
[--t0 T0] [--kf KF] [--kb KB] [-a ATOL] [-r RTOL]
[--logy] [--logt] [--num-jac] [--plot]
[--savefig SAVEFIG] [--splitplots] [--plotlogy]
[--plotsymlogy] [--plotlogt] [--scale-err SCALE_ERR]
[--scaling SCALING] [-v]
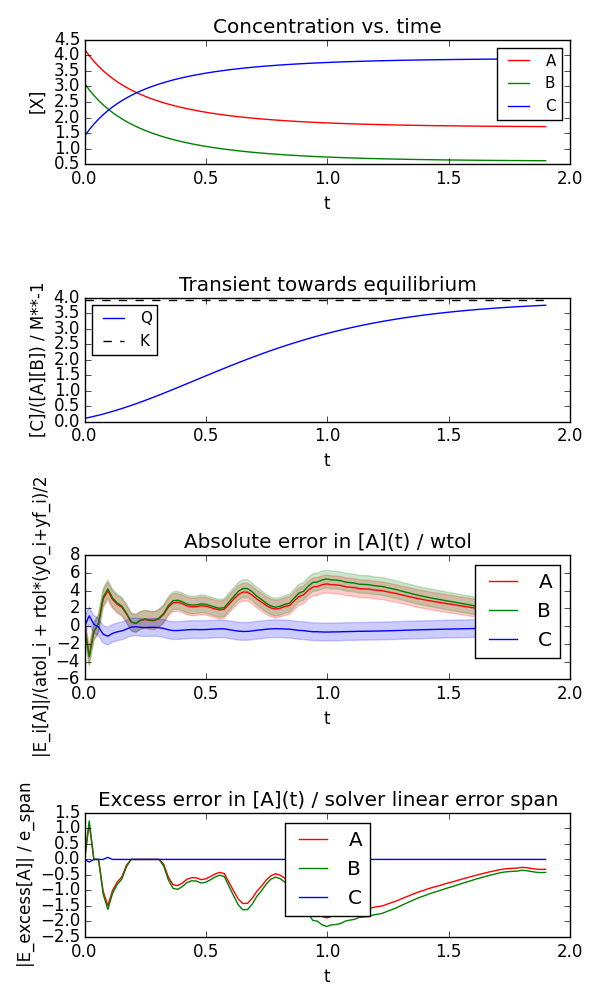
Runs the integration and (optionally) plots:
- Individual concentrations as function of time
- Reaction Quotient vs. time (with equilibrium constant as reference)
- Numerical error commited (with tolerance span plotted)
- Excess error committed (deviation outside tolerance span)
Concentrations (A0, B0, C0) are taken to be in "M" (molar),
kf in "M**-1 s**-1" and kb in "s**-1", t0 and tend in "s"
optional arguments:
-h, --help show this help message and exit
--tend TEND 1.9
-A A0, --A0 A0 4.2
-B B0, --B0 B0 3.1
-C C0, --C0 C0 1.4
--nt NT 100
--t0 T0 0.0
--kf KF 0.9
--kb KB 0.23
-a ATOL, --atol ATOL u'1e-7,1e-6,1e-5'
-r RTOL, --rtol RTOL u'1e-6'
--logy False
--logt False
--num-jac False
--plot False
--savefig SAVEFIG u'None'
--splitplots False
--plotlogy False
--plotsymlogy False
--plotlogt False
--scale-err SCALE_ERR
1.0
--scaling SCALING 1.0
-v, --verbose False
Here is an example generated by:
$ python equilibrium.py --plot --savefig equilibrium.png
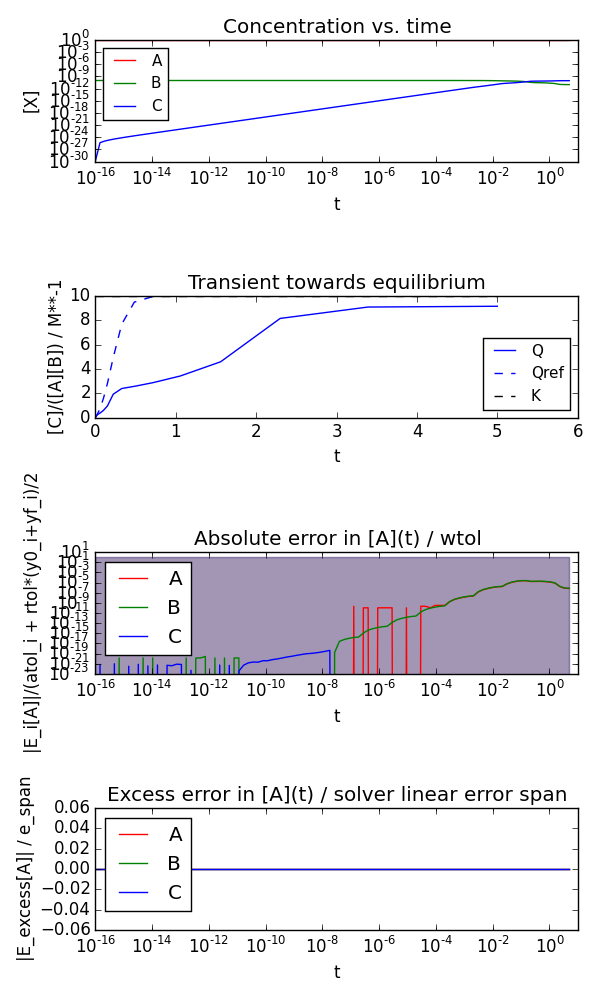
If concentrations are far from 1 (and below abstol) the accuracy of the numerical solution will be very poor:
$ python equilibrium.py --A0 1.0 --B0 1e-10 --C0 1e-30 --kf 10 --kb 1 --t0 0 --tend 5 --plot --plotlogy --plotlogt --savefig equilibrium_unscaled.png
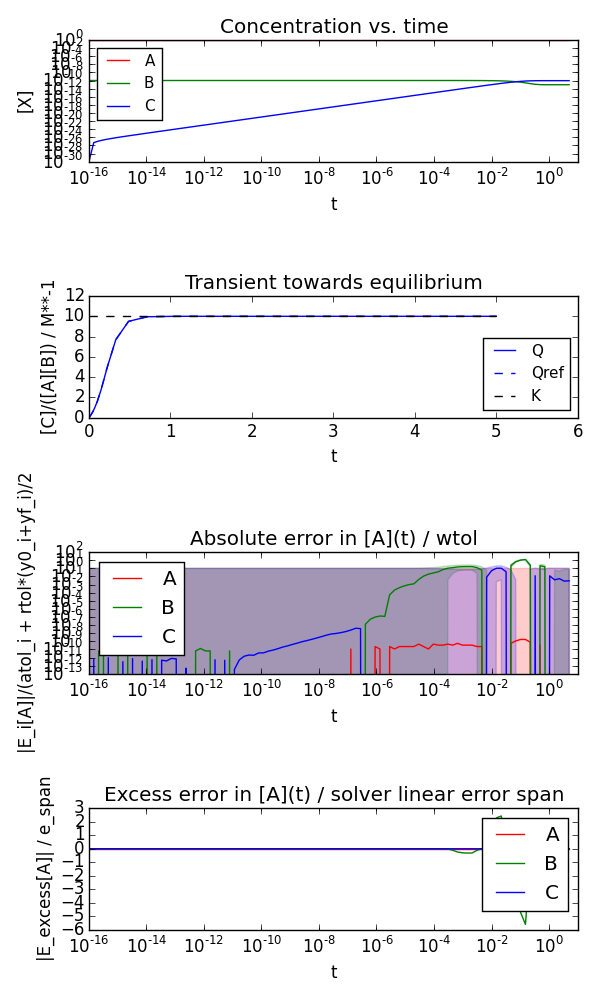
But by scaling the concentrations so that the smallest is well above the absolute tolerance we can get accurate results:
$ python equilibrium.py --scaling 1e10 --A0 1.0 --B0 1e-10 --C0 1e-30 --kf 10 --kb 1 --t0 0 --tend 5 --plot --plotlogy --plotlogt --savefig equilibrium_scaled.png
- equilibrium.analytic_x(t, A, B, C, kf, kb, _exp=<ufunc 'exp'>)[source]¶
- Analytic solution to the dimeriztion reaction:
- A + B <=> C; (K = kf/kb)
- equilibrium.integrate_rd(tend=1.9, A0=4.2, B0=3.1, C0=1.4, nt=100, t0=0.0, kf=0.9, kb=0.23, atol=u'1e-7, 1e-6, 1e-5', rtol=u'1e-6', logy=False, logt=False, num_jac=False, plot=False, savefig=u'None', splitplots=False, plotlogy=False, plotsymlogy=False, plotlogt=False, scale_err=1.0, scaling=1.0, verbose=False)[source]¶
Runs the integration and (optionally) plots:
- Individual concentrations as function of time
- Reaction Quotient vs. time (with equilibrium constant as reference)
- Numerical error commited (with tolerance span plotted)
- Excess error committed (deviation outside tolerance span)
Concentrations (A0, B0, C0) are taken to be in “M” (molar), kf in “M**-1 s**-1” and kb in “s**-1”, t0 and tend in “s”